FOR-COVID
Bavarian consortium for research on the pandemic disease COVID-19 (FOR-COVID)

Deciphering SARS-CoV-2 infection by scSLAM-seq and artificial intelligence
Projectleaders
Prof. Dr. Lars Dölken
Dr. Florian Erhard
Dr. Antoine-Emmanuel Saliba
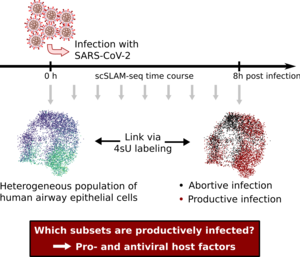
Upon virus entry into a cell, the cell either manages to control the virus or succumbs to it. The mechanisms underlying this heterogeneity remain elusive. We recently introduced single-cell, thiol-(SH)-linked alkylation of RNA for metabolic labelling sequencing (scSLAM-seq) to decipher host-pathogen interaction with unprecedented temporal resolution at single cell level. During the initial FOR-COVID funding phase, we successfully improved on our scSLAM-seq approach that is now amenable to profile thousands of single-cells. Furthermore, we developed the computational analysis pipeline GRAND-SLAM 3.0 tailored to handle large-scale scSLAMseq data obtained by droplet-based 10x Chromium single cell RNA-seq. We recorded the transcriptional responses in >3,000 human epithelial cells infected with SARS-CoV-2 during the first 8 h of infection with 2 h sliding sampling windows. Our data indicate that rapid transcriptional responses, which are only detectable in “new RNA” during the first two hours of infection, enable clear predictions on infection outcome at 8 h post infection. Based on more extensive scSLAM-seq data on SARS-CoV-2 infection of primary human alveolar epithelial cells, we now aim to identify novel cellular pro- and antiviral factors that define infection outcome. We will compare early cellular response to SARS-CoV-2 infections with two other important respiratory viruses, namely influenza A virus and respiratory syncytial virus. We will interconnect cell-state transitions at single cell level over consecutive time windows to dissect how the state of a given cell at the time of infection defines infection outcome. A long-term goal is to develop artificial intelligence (AI) systems to make testable predictions about how specific cell types and states react to SARS-CoV-2 and other incoming viruses in terms of their gene expression programs.
Project partners:
- Helmholtz Institute for RNA based Infection Research (HIRI) Würzburg
- Julius-Maximilians-Universität of Würzburg



